DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome | eLife
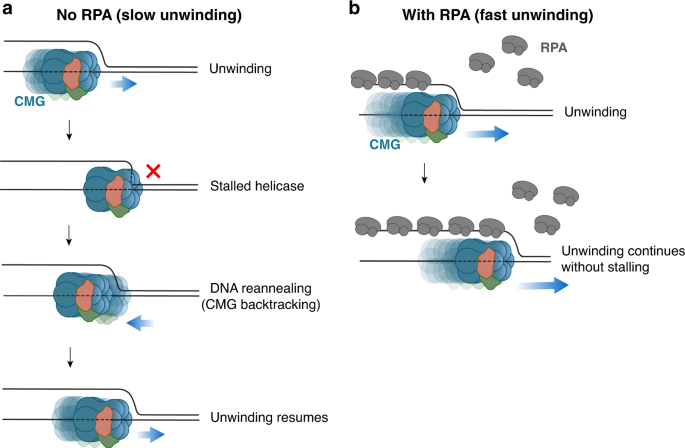
Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase | Nature Communications
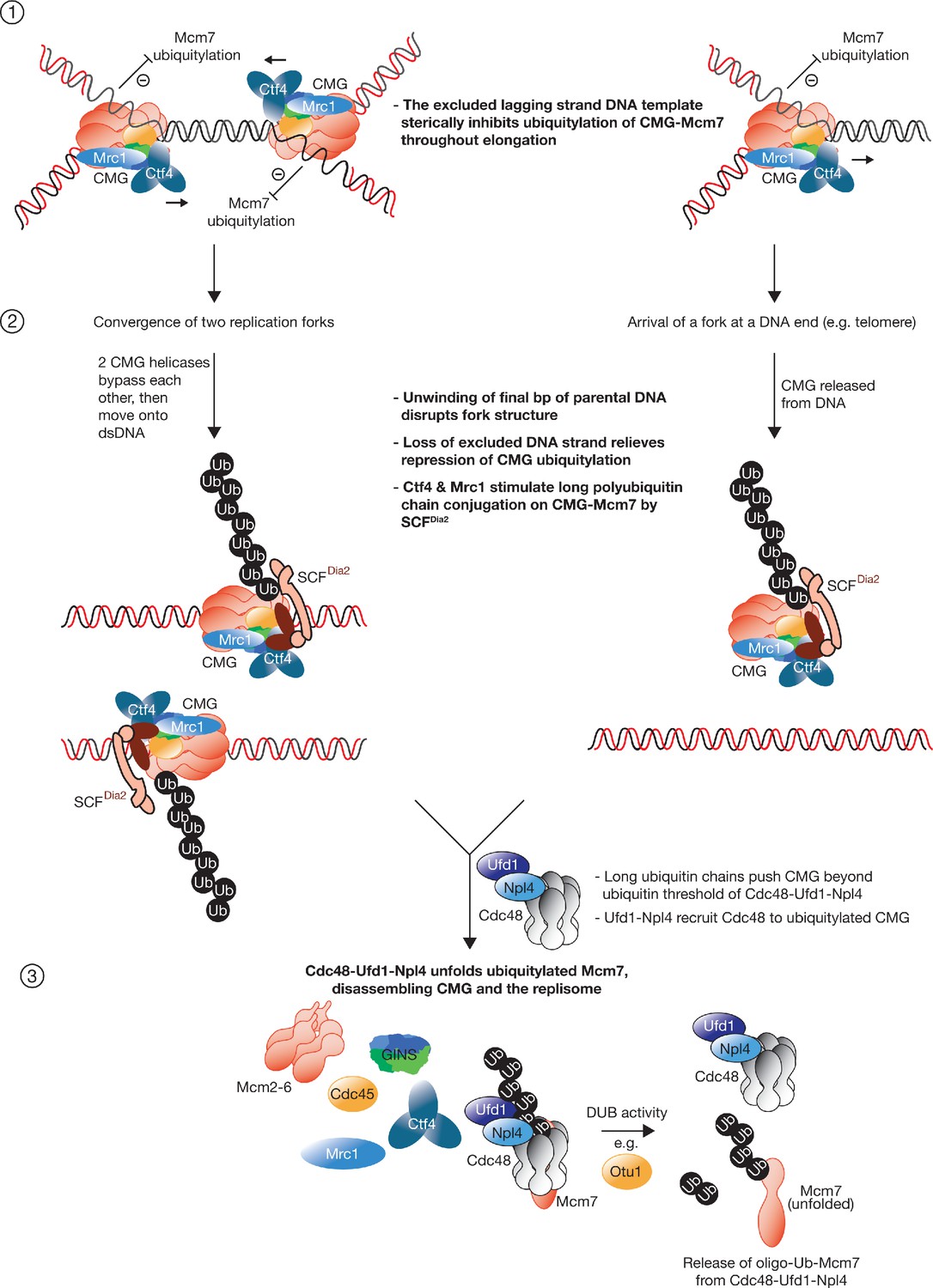
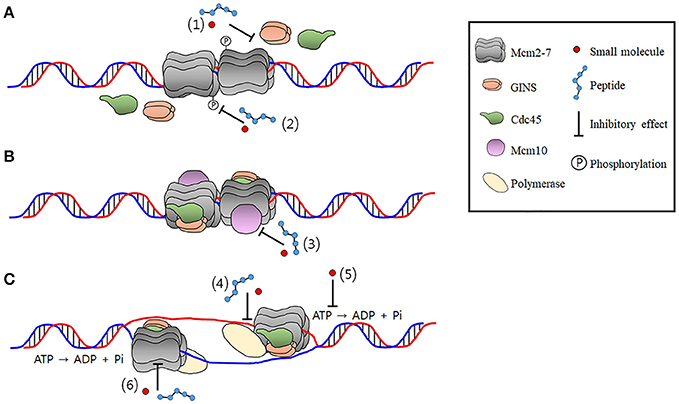
CMG helicase disassembly is controlled by replication fork DNA, replisome components and a ubiquitin threshold | eLife

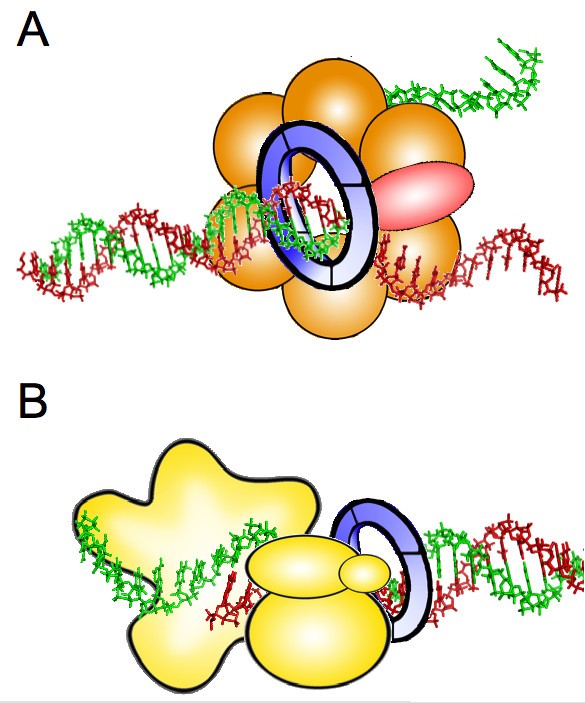
Schematic structure of the CMG helicase. (A) Two transloaction models... | Download Scientific Diagram
![PDF] The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication | Semantic Scholar PDF] The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/2bed3134525627f77020b000c771139c73709545/8-Figure7-1.png)
PDF] The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication | Semantic Scholar

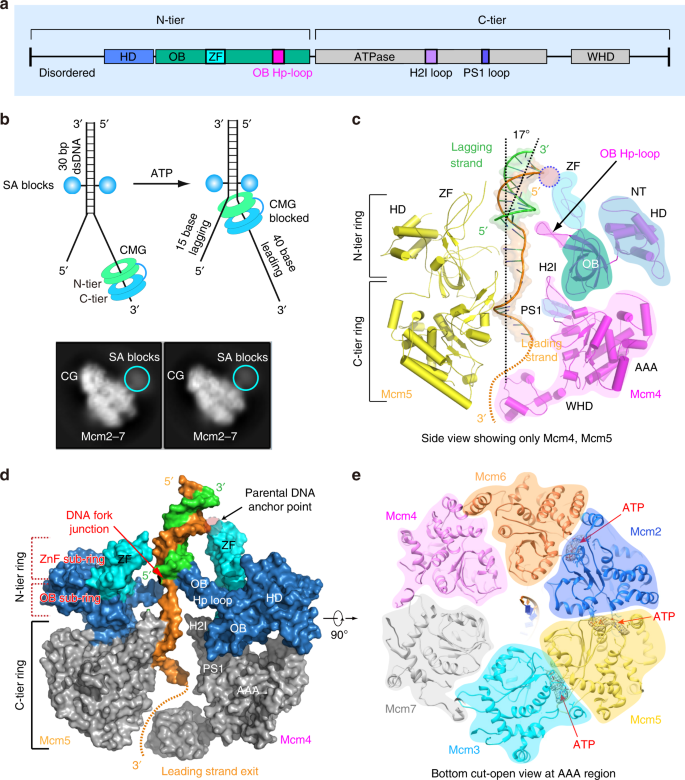
Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation | PNAS

Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation | PNAS

Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication | Science

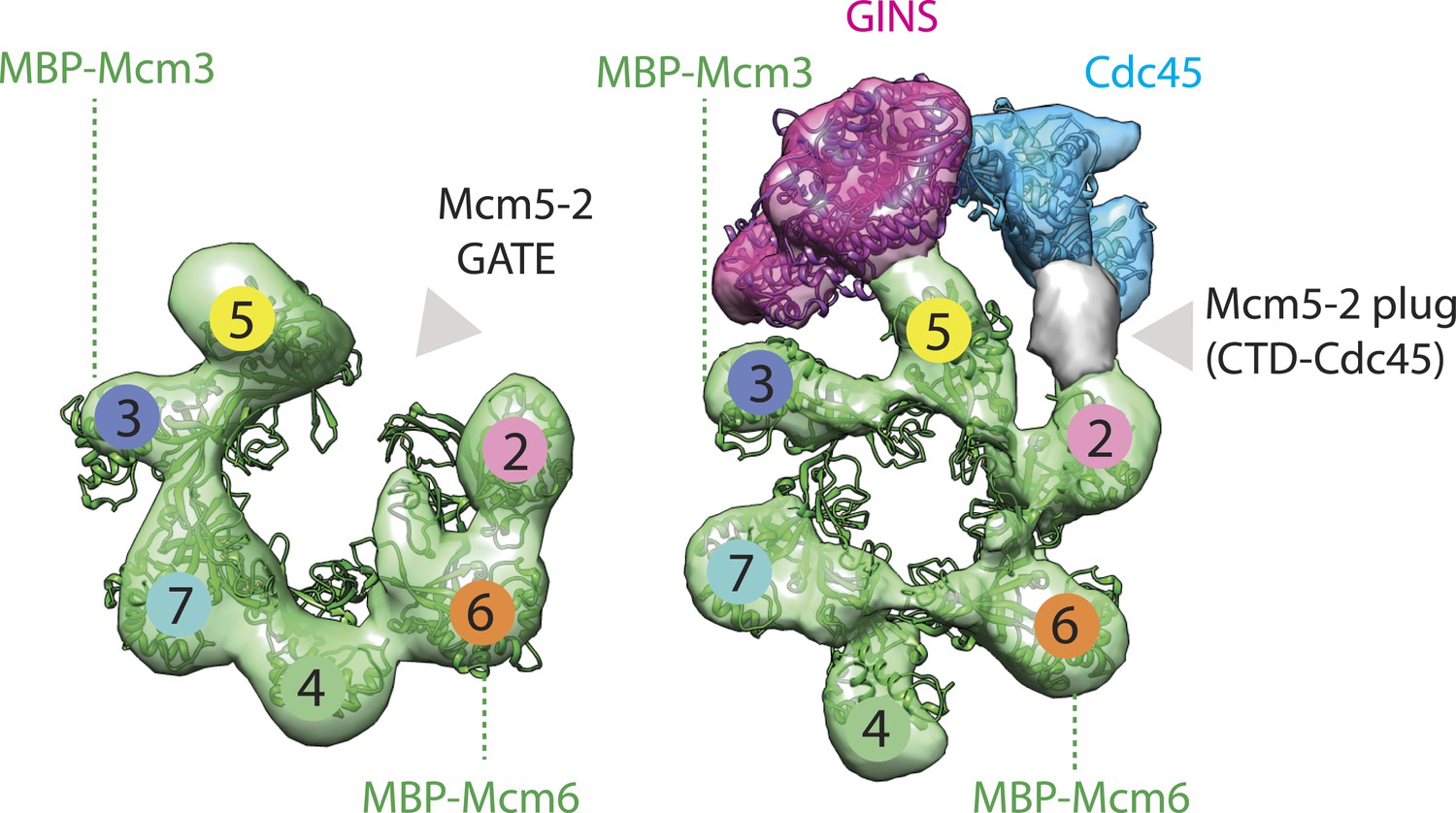
Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase - ScienceDirect

CMG helicase structure and translocation mechanism. (a) Low resolution... | Download Scientific Diagram
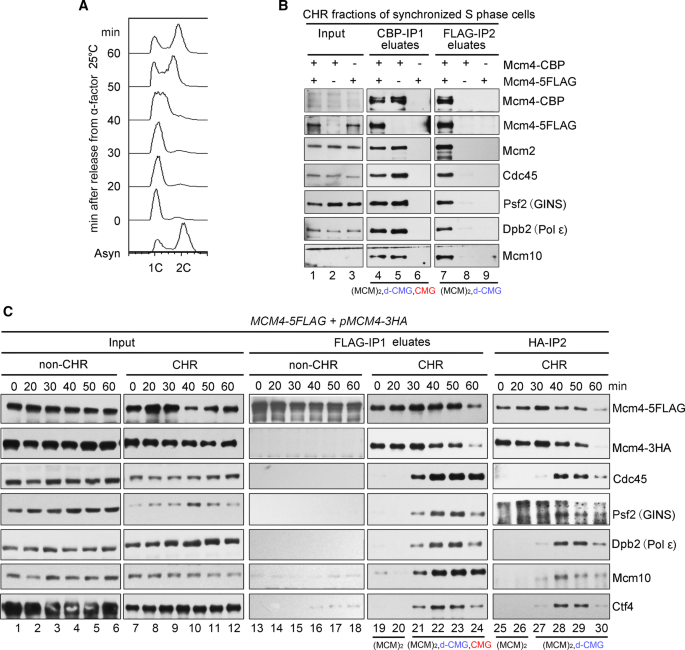
Characterization of the dimeric CMG/pre-initiation complex and its transition into DNA replication forks | SpringerLink

Spotlight on the Replisome: Aetiology of DNA Replication-Associated Genetic Diseases: Trends in Genetics

Mcm10 regulates the assembly of the CMG complex and DNA origin melting.... | Download Scientific Diagram

Pre‐initiation complex assembly functions as a molecular switch that splits the Mcm2‐7 double hexamer | EMBO reports
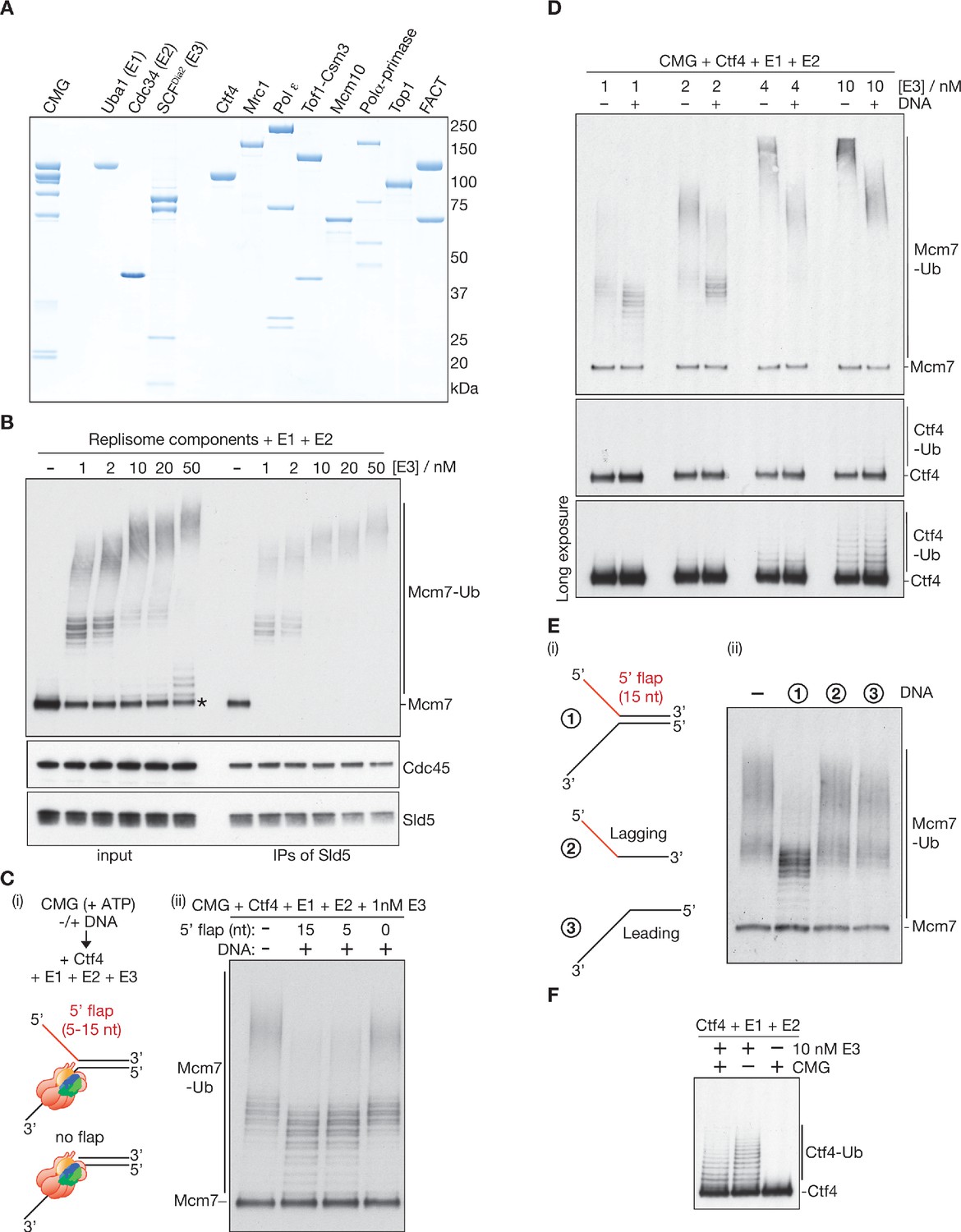
CMG helicase disassembly is controlled by replication fork DNA, replisome components and a ubiquitin threshold | eLife

Mechanisms in helicase activation and analysis of the replication fork activity and architecture – Speck Lab / DNA Replication Group – Christian Speck
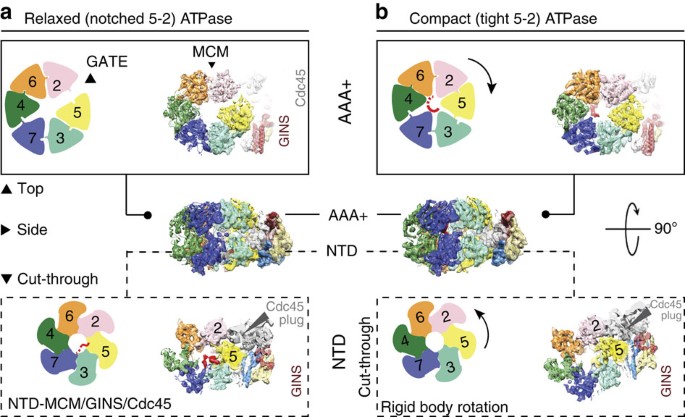
Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate | Nature Communications

CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication | PNAS
Isolation of the Cdc45 Mcm2–7 GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase